# \donttest{
library(shinymrp)
# Initialize the MRP workflow
workflow <- mrp_workflow()
# Load example data
sample_data <- example_sample_data()
### DATA PREPARATION
# Preprocess sample data
workflow$preprocess(
sample_data,
is_timevar = TRUE,
is_aggregated = TRUE,
special_case = NULL,
family = "binomial"
)
#> Preprocessing sample data...
# Link data to the ACS
# and obtain poststratification data
workflow$link_acs(
link_geo = "zip",
acs_year = 2021
)
#> Linking data to the ACS...
### DESCRIPTIVE STATISTICS
# Visualize demographic distribution of data
sex_bars <- workflow$demo_bars(demo = "sex")
# Visualize geographic distribution of data
ss_map <- workflow$sample_size_map()
#> Registered S3 method overwritten by 'quantmod':
#> method from
#> as.zoo.data.frame zoo
# Visualize outcome measure
raw_outcome_plot <- workflow$outcome_plot()
### MODEL BUILDING
# Create new model objects
model <- workflow$create_model(
intercept_prior = "normal(0, 4)",
fixed = list(
sex = "normal(0, 2)",
race = "normal(0, 2)"
),
varying = list(
age = "",
time = ""
)
)
# Run MCMC
model$fit(n_iter = 500, n_chains = 2, seed = 123)
#> Running MCMC with 2 parallel chains, with 1 thread(s) per chain...
#>
#> Chain 1 Iteration: 1 / 500 [ 0%] (Warmup)
#> Chain 2 Iteration: 1 / 500 [ 0%] (Warmup)
#> Chain 1 Iteration: 50 / 500 [ 10%] (Warmup)
#> Chain 2 Iteration: 50 / 500 [ 10%] (Warmup)
#> Chain 1 Iteration: 100 / 500 [ 20%] (Warmup)
#> Chain 2 Iteration: 100 / 500 [ 20%] (Warmup)
#> Chain 1 Iteration: 150 / 500 [ 30%] (Warmup)
#> Chain 2 Iteration: 150 / 500 [ 30%] (Warmup)
#> Chain 1 Iteration: 200 / 500 [ 40%] (Warmup)
#> Chain 2 Iteration: 200 / 500 [ 40%] (Warmup)
#> Chain 1 Iteration: 250 / 500 [ 50%] (Warmup)
#> Chain 1 Iteration: 251 / 500 [ 50%] (Sampling)
#> Chain 2 Iteration: 250 / 500 [ 50%] (Warmup)
#> Chain 2 Iteration: 251 / 500 [ 50%] (Sampling)
#> Chain 1 Iteration: 300 / 500 [ 60%] (Sampling)
#> Chain 2 Iteration: 300 / 500 [ 60%] (Sampling)
#> Chain 1 Iteration: 350 / 500 [ 70%] (Sampling)
#> Chain 2 Iteration: 350 / 500 [ 70%] (Sampling)
#> Chain 1 Iteration: 400 / 500 [ 80%] (Sampling)
#> Chain 1 Iteration: 450 / 500 [ 90%] (Sampling)
#> Chain 2 Iteration: 400 / 500 [ 80%] (Sampling)
#> Chain 2 Iteration: 450 / 500 [ 90%] (Sampling)
#> Chain 1 Iteration: 500 / 500 [100%] (Sampling)
#> Chain 2 Iteration: 500 / 500 [100%] (Sampling)
#> Chain 1 finished in 1.1 seconds.
#> Chain 2 finished in 1.1 seconds.
#>
#> Both chains finished successfully.
#> Mean chain execution time: 1.1 seconds.
#> Total execution time: 1.2 seconds.
#>
#> Warning: 1 of 500 (0.0%) transitions ended with a divergence.
#> See https://mc-stan.org/misc/warnings for details.
# Estimates summary and diagnostics
model$summary()
#> $fixed
#> Estimate Est.Error l-95% CI u-95% CI R-hat Bulk_ESS
#> intercept -0.1552038 0.2029703 -0.51179360 0.2636422 1.0015135 155.1940
#> sex.male 0.2824838 0.1338178 0.02599781 0.5476556 0.9973944 621.5216
#> race.black NA NA NA NA NA NA
#> race.other 0.7682242 0.1672083 0.45840460 1.0993380 0.9987605 456.4001
#> race.white 0.8251199 0.1669716 0.50039767 1.1567253 0.9999359 348.4526
#> Tail_ESS
#> intercept 148.2918
#> sex.male 421.2527
#> race.black NA
#> race.other 366.2108
#> race.white 395.3222
#>
#> $varying
#> Estimate Est.Error l-95% CI u-95% CI R-hat Bulk_ESS
#> age (intercept) 0.2687213 0.1887081 0.02469051 0.7533799 1.021851 123.6085
#> time (intercept) 0.2716189 0.1202040 0.07451838 0.5388200 1.005514 159.8250
#> Tail_ESS
#> age (intercept) 192.4378
#> time (intercept) 159.7993
#>
#> $residual
#> data frame with 0 columns and 0 rows
#>
#> $spatial_proportion
#> data frame with 0 columns and 0 rows
#>
# Sampling diagnostics
model$diagnostics()
#> Metric
#> 1 Divergence
#> 2 Maximum tree depth
#> 3 E-BFMI
#> Message
#> 1 1 of 500 (0.2%) transitions ended with a divergence
#> 2 0 of 500 transitions hit the maximum tree depth limit of 10
#> 3 0 of 2 chains had an E-BFMI less than 0.3
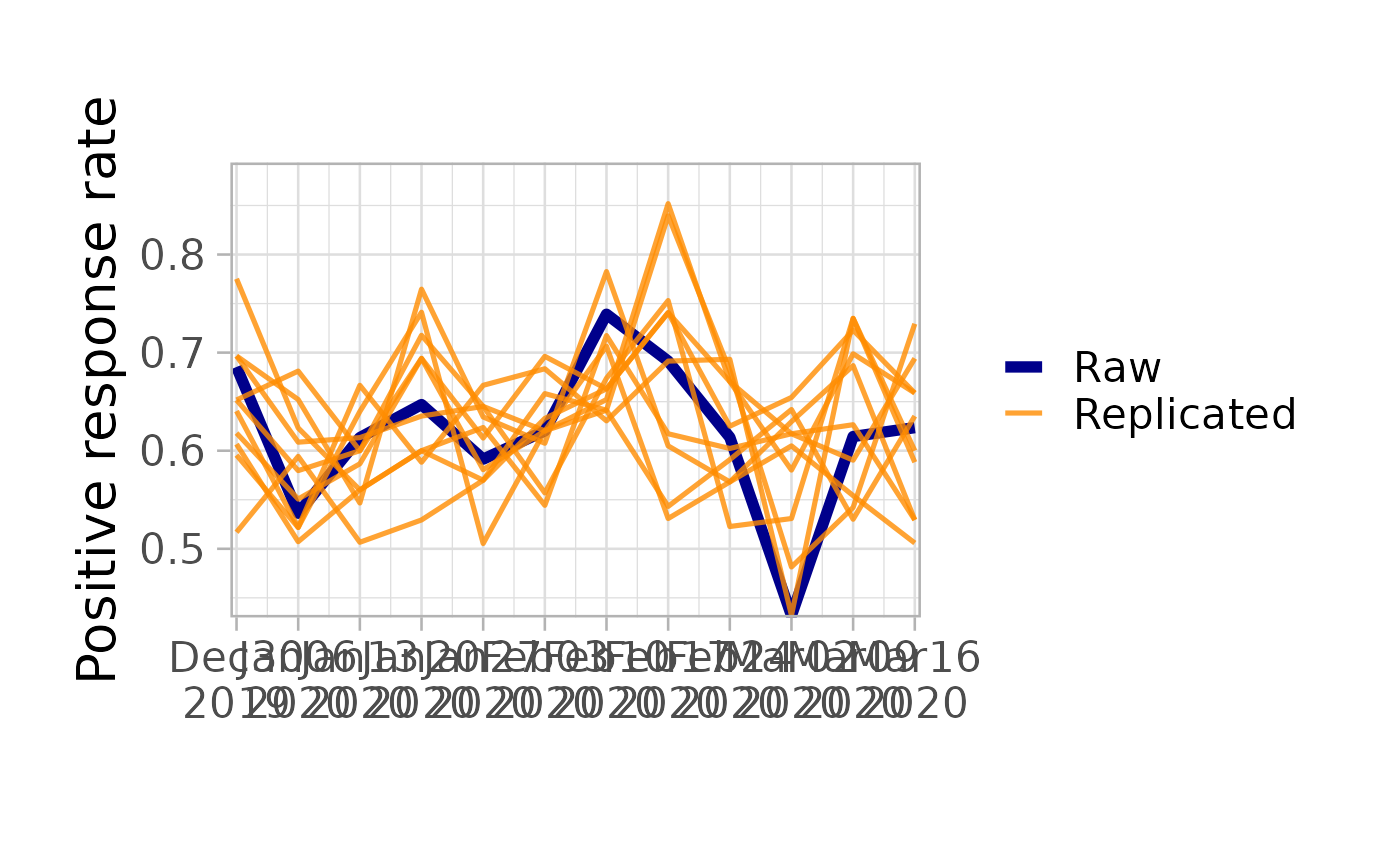
# Posterior predictive check
workflow$pp_check(model)
#> Running posterior predictive check...
 ### VISUALIZE RESULTS
# Plots of overall estimates, estimates for demographic groups, and geographic areas
workflow$estimate_plot(model, group = "sex")
#> Running poststratification...
### VISUALIZE RESULTS
# Plots of overall estimates, estimates for demographic groups, and geographic areas
workflow$estimate_plot(model, group = "sex")
#> Running poststratification...
 # Choropleth map of estimates for geographic areas
workflow$estimate_map(model, geo = "county")
# }
# Choropleth map of estimates for geographic areas
workflow$estimate_map(model, geo = "county")
# }
 ### VISUALIZE RESULTS
# Plots of overall estimates, estimates for demographic groups, and geographic areas
workflow$estimate_plot(model, group = "sex")
#> Running poststratification...
### VISUALIZE RESULTS
# Plots of overall estimates, estimates for demographic groups, and geographic areas
workflow$estimate_plot(model, group = "sex")
#> Running poststratification...
 # Choropleth map of estimates for geographic areas
workflow$estimate_map(model, geo = "county")
# }
# Choropleth map of estimates for geographic areas
workflow$estimate_map(model, geo = "county")
# }